Журнал "Гастроэнтерология" Том 59, №3, 2025
Вернуться к номеру
Порушення смаку в дітей з ожирінням: генетичні аспекти рецепції
Авторы: O.E. Abaturov, A.O. Nikulina
Dnipro State Medical University, Dnipro, Ukraine
Рубрики: Гастроэнтерология
Разделы: Клинические исследования
Версия для печати
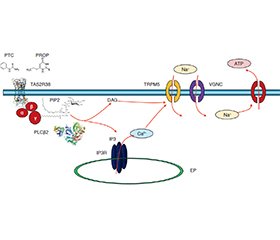
Актуальність. Численні однонуклеотидні варіанти (single nucleotide variants — SNV) гена члена 38 рецептора смаку 2 (taste 2 receptor member 38 — TAS2R38) зумовлюють формування індивідуальних особливостей сприйняття гіркого смаку. Мета: дослідити асоціації SNV rs10246939, rs1726866, rs713598 гена TAS2R38 із ризиком розвитку метаболічно нездорового ожиріння (metabolically unhealthy obesity — MUO) у дітей. Матеріали та методи. Обстежено 400 дітей віком 6–18 років, з яких проліковано 350 пацієнтів з ожирінням. Контрольну групу становили 50 дітей без ожиріння. Серед пацієнтів з ожирінням за рекомендаціями консорціуму IDEFICS були сформовані дві підгрупи: з MUO (n = 204) та метаболічно здоровим ожирінням (metabolically healthy obesity — MHO) (n = 146). Рівень смакових уподобань визначали за опитувальником FBPQ. Базальну глікемію, інсулінемію досліджували імунохімічним методом з електрохемілюмінесцентною детекцією, уміст ліпопротеїнів високої щільності й тригліцеридів — ферментативно-колориметричним методом в лабораторії Synevo (Дніпро, Україна). SNV гена TAS2R38 ідентифікували повногеномним секвенуванням наступного покоління в 52 пацієнтів (31 — із MUO та 21 — із MHO) в лабораторії CeGat (Тюбінген, Німеччина). Результати. Середні рівні (М ± m) смакових уподобань у групах порівняння за опитувальником FBPQ вірогідно відрізнялись до солодкого (у дітей з ожирінням — (3,36 ± 0,08) бала, у групі контролю — (3,74 ± 0,07) бала, p < 0,002) та гіркого смаків (у дітей з ожирінням становили (2,77 ± 0,15) бала, у контрольній групі — (3,37 ± 0,15) бала, p < 0,00013). Середні рівні смакових уподобань у дітей із MUO порівнянo з дітьми з MHO вірогідно відрізнялись до гіркого смаку — відповідно (2,75 ± 0,12) бала проти (3,24 ± 0,05) бала, p < 0,02. Ідентифіковано чотири SNV гена TAS2R38: rs713598, rs1726866, rs10246939, rs145970530. Висновки. Генотип CG rs713598 гена TAS2R38 пов’язаний із підвищеним ризиком розвитку MUO та кардіометаболічних порушень.
Background. Numerous single nucleotide variants (SNV) of the taste 2 receptor member 38 (TAS2R38) gene determine the formation of individual characteristics of bitter taste perception. The аim: to study the association of SNV rs10246939, rs1726866, rs713598 of the TAS2R38 gene with the risk of metabolically unhealthy obesity (MUO) in children. Materials and methods. Four hundred children aged 6–18 years were examined, of which 350 with obesity were treated. The control group was represented by 50 children without obesity. Among obese children, two observation subgroups were formed: MUO (n = 204) and metabolically healthy obesity (MHO) (n = 146). The level of taste preferences was determined by the FBPQ. The level of basal glycemia, insulinemia was studied by immunochemical method with electro chemiluminescent detection, high-density lipoproteins and triglycerides — by enzymatic-colorimetric method in the Synevo (Ukraine). SNVs of the TAS2R38 gene were identified by whole-genome next-generation sequencing in 52 patients at the CeGat laboratory (Germany). Results. The mean levels (M ± m) of taste preferences in the comparison groups according to the FBPQ were significantly different for sweet (in obese children — (3.36 ± 0.08) points, while in the control group (3.74 ± 0.07) points, p < 0.002) and bitter tastes (in obese children it was (2.77 ± 0.15) points, while in the control group (3.37 ± 0.15) points, p < 0.00013). The mean levels of taste preferences in children with MUO compared to those with MHO were significantly different for bitter tastes — (2.75 ± 0.12) points versus (3.24 ± 0.05) points, respectively, p < 0.02. We identified four SNVs in the TAS2R38 gene: rs713598, rs1726866, rs10246939, rs145970530. Conclusions. CG genotype of rs713598 in the TAS2R38 gene is associated with an increased risk of developing MUO and cardiometabolic disorders.
діти; ожиріння; однонуклеотидні варіанти; член 38 рецептора смаку 2
children; obesity; single nucleotide variants; taste 2 receptor member 38
Для ознакомления с полным содержанием статьи необходимо оформить подписку на журнал.
- Taruno A, Nomura K, Kusakizako T, et al. Taste transduction and channel synapses in taste buds. Pflugers Arch. 2021 Jan;473(1):3-13. doi: 10.1007/s00424-020-02464-4.
- Barlow LA. The sense of taste: Development, regeneration, and dysfunction. WIREs Mech Dis. 2022 May;14(3):e1547. doi: 10.1002/wsbm.1547.
- Wilson CE, Lasher RS, Yang R, et al. Taste Bud Connectome: Implications for Taste Information Processing. J Neurosci. 2022 Feb 2;42(5):804-816. doi: 10.1523/JNEUROSCI.0838-21.2021.
- Ahmad R, Dalziel JEG. Protein-Coupled Receptors in Taste Physiology and Pharmacology. Front Pharmacol. 2020 Nov 30;11:587664. doi: 10.3389/fphar.2020.587664.
- Nishihara H, Toda Y, Kuramoto T, et al. A vertebrate-wide ca–talogue of T1R receptors reveals diversity in taste perception. Nat Ecol Evol. 2024 Jan;8(1):111-120. doi: 10.1038/s41559-023-02258-8.
- Wooding SP, Ramirez VA, Behrens M. Bitter taste receptors: Genes, evolution and health. Evol Med Public Health. 2021 Oct 13;9(1):431-447. doi: 10.1093/emph/eoab031.
- Descamps-Solà M, Vilalta A, Jalsevac F, et al. Bitter taste receptors along the gastrointestinal tract: comparison between humans and rodents. Front Nutr. 2023 Aug 30;10:1215889. doi: 10.3389/fnut.2023.1215889.
- Smail HO. The roles of genes in the bitter taste. AIMS Genet. 2019 Dec 24;6(4):88-97. doi: 10.3934/genet.2019.4.88.
- Jeruzal-Świątecka J, Fendler W, Pietruszewska W. Clinical Role of Extraoral Bitter Taste Receptors. Int J Mol Sci. 2020 Jul 21;21(14):5156. doi: 10.3390/ijms21145156.
- Itoigawa A, Toda Y, Kuraku S, et al. Evolutionary origins of bitter taste receptors in jawed vertebrates. Curr Biol. 2024 Apr 8;34(7):R271-R272. doi: 10.1016/j.cub.2024.02.024.
- Zhao J, Yin J, Wang Z, et al. Complicated gene network for regulating feeding behavior: novel efficient target for pest management. Pest Manag Sci. 2025 Jan;81(1):10-21. doi: 10.1002/ps.8459.
- Abaturov A, Nikulina A. Taste preferences and obesity. Pediatria Polska — Polish Journal of Paediatrics. 2022;97(1):1-6. doi: 10.5114/polp.2022.115139.
- Chamoun E, Carroll NA, Duizer LM, et al. Guelph Family Health Study. The Relationship between Single Nucleotide Polymorphisms in Taste Receptor Genes, Taste Function and Dietary Intake in Preschool-Aged Children and Adults in the Guelph Family Health Study. Nutrients. 2018 Jul 29;10(8):990. doi: 10.3390/nu10080990.
- Robino A, Rosso N, Guerra M, et al. Taste perception and expression in stomach of bitter taste receptor tas2r38 in obese and lean subjects. Appetite. 2021 Nov 1;166:105595. doi: 10.1016/j.appet.2021.105595.
- Ramos-Lopez O, Martinez-Aceviz Y, Sobrevilla-Navarro AA, et al. Genetic Influence on Capsaicin Tolerance: Precision Nutrition Implications for Obesity Handling. Lifestyle Genom. 2024;17(1):57-63. doi: 10.1159/000539293.
- Itoigawa A, Nakagita T, Toda Y. The Remarkable Diversity of Vertebrate Bitter Taste Receptors: Recent Advances in Genomic and Functional Studies. Int J Mol Sci. 2024 Nov 25;25(23):12654. doi: 10.3390/ijms252312654.
- Zafirovska M, Zafirovski A, Režen T, et al. The Outcome of Metabolic and Bariatric Surgery in Morbidly Obese Patients with Different Genetic Variants Associated with Obesity: A Systematic Review. Nutrients. 2024 Aug 1;16(15):2510. doi: 10.3390/nu16152510.
- Coltell O, Sorlí JV, Asensio EM, et al. Association between taste perception and adiposity in overweight or obese older subjects with metabolic syndrome and identification of novel taste-related genes. Am J Clin Nutr. 2019 Jun 1;109(6):1709-1723. doi: 10.1093/ajcn/nqz038.
- Jo YS, Choi JH. Genetic variation in TAS2R38 bitterness receptor is associated with body composition in Korean females. Int J Food Sci Nutr. 2024 Mar;75(2):197-206. doi: 10.1080/09637486.2023.2294682.
- Shushari MK, Wei T, Tapanee P, et al. The Influence of Taste Genes on Body Fat and Alcohol Consumption. Nutrients. 2024 Jun 4;16(11):1756. doi: 10.3390/nu16111756.
- Little J, Higgins JP, Ioannidis JP, et al. STrengthening the REporting of Genetic Association Studies (STREGA): An Extension of the STROBE Statement. Genet Epidemiol. 2009 Nov;33(7):581-98. doi: 10.1002/gepi.20410.
- Alberti KG, Zimmet P, Kaufman F, et al. The IDF consensus definition of the metabolic syndrome in children and adolescents. International Diabetes Federation. 2017:17-19. Available from: https://www.idf.org/e-library/consensus-statements/61-idf-consensus-definition-of-metabolic-syndrome-in-children-and-adolescents.
- Flynn JT, Kaelber DC, Baker-Smith CM, et al. Subcommittee on screening and management of high blood pressure in children. Clinical Practice Guideline for Screening and Management of High Blood Pressure in Children and Adolescents. Pediatrics. 2017 Sep;140(3):e20171904. doi: 10.1542/peds.2017-1904.
- De Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bulletin of the World Health Organization. 2007;85:660-7. Available from: http://www.who.int/growthref/growthref_who_bull/en/index.html.
- Draznin B, Aroda VR, Bakris G, et al. American Diabetes Association Professional Practice Committee. 6. Glycemic targets: Standards of Medical Care in Diabetes — 2022. Diabetes Care 2022;45(Suppl 1):83-96. doi: 10.2337/dc22-S006.
- Peplies J, Börnhorst C, Günther K, et al. IDEFICS consortium. Longitudinal associations of lifestyle factors and weight status with insulin resistance (HOMA-IR) in preadolescent children: the large prospective cohort study IDEFICS. Int J Behav Nutr Phys Act. 2016 Sep 2;13(1):97. doi: 10.1186/s12966-016-0424-4.
- Elkins C, Fruh Sh, Jones L, et al. Clinical Practice Recommendations for Pediatric Dyslipidemia. Journal of Pediatric Health Care. 2019;33(4):494-504. doi: 10.1016/j.pedhc.2019.02.009.
- Jilani HS, Intemann T, Bogl LH, et al. Familial aggregation and socio-demographic correlates of taste preferences in European children. BMC Nutr. 2017;3:87. doi: 10.1186/s40795-017-0206-7.
- Zhang J, Yao Y, He H, et al. Clinical Interpretation of Sequence Variants. Curr Protoc Hum Genet. 2020;106(1):e98. doi: 10.1002/cphg.98.
- Liao X, Li M, Zou Y, et al. An Efficient Trimming Algorithm based on Multi-Feature Fusion Scoring Model for NGS Data. IEEE/ACM Trans Comput Biol Bioinform. 2020;17(3):728-738. doi: 10.1109/TCBB.2019.2897558.
- Gunning AC, Fryer V, Fasham J, et al. Assessing performance of pathogenicity predictors using clinically relevant variant datasets. J Med Genet. 2021 Aug;58(8):547-555. doi: 10.1136/jmedge–net-2020-107003.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics. 2009;25(14):1754-1760. doi: 10.1093/bioinformatics/btp324.
- Livesey BJ, Marsh JA. Using deep mutational scanning to benchmark variant effect predictors and identify disease mutations. Mol Syst Biol. 2020 Jul;16(7):e9380. doi: 10.15252/msb.20199380.
- Mahmood K, Jung CH, Philip G, et al. Variant effect prediction tools assessed using independent, functional assay-based datasets: implications for discovery and diagnostics. Hum Genomics. 2017 May 16;11(1):10. doi: 10.1186/s40246-017-0104-8.
- Deelen P, Bonder MJ, van der Velde KJ, et al. Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res Notes. 2014;7:901. doi: 10.1186/1756-0500-7-901.
- Mose LE, Wilkerson MD, Hayes DN, et al. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics. 2014;30(19):2813-2815. doi: 10.1093/bioinformatics/btu376.
- RefSeq: NCBI Reference Sequence Database. Available from: https://www.ncbi.nlm.nih.gov/refseq.
- Khan AS, Murtaza B, Hichami A, et al. A cross-talk between fat and bitter taste modalities. Biochimie. 2019 Apr;159:3-8. doi: 10.1016/j.biochi.2018.06.013.
- Wang X, Wang L, Xia M, et al. Variations in the TAS2R38 gene among college students in Hubei. Hereditas. 2022 Dec 19;159(1):46. doi: 10.1186/s41065-022-00260-x.
- Tuzim K, Korolczuk A. An update on extra-oral bitter taste receptors. J Transl Med. 2021 Oct 21;19(1):440. doi: 10.1186/s12967-021-03067-y.
- Shivam V. A meta-analysis on polymorphic trait of taste perception mediated by TAS2R38 genotype. Exp Clin Psychopharmacol. 2024 Oct;32(5):497-505. doi: 10.1037/pha0000728.
- Miguet L, Zhang Z, Grigorov MG. Computational studies of ligand-receptor interactions in bitter taste receptors. J Recept Signal Transduct Res. 2006;26(5–6):611-30. doi: 10.1080/10799890600928210.
- Mao Z, Cheng W, Li Z, et al. Clinical Associations of Bitter Taste Perception and Bitter Taste Receptor Variants and the Potential for Personalized Healthcare. Pharmgenomics Pers Med. 2023 Feb 12;16:121-132. doi: 10.2147/PGPM.S390201.
- Perna S, Riva A, Nicosanti G, et al. Association of the bitter taste receptor gene TAS2R38 (polymorphism RS713598) with sensory responsiveness, food preferences, biochemical parameters and body-composition markers. A cross-sectional study in Italy. Int J Food Sci Nutr. 2018 Mar;69(2):245-252. doi: 10.1080/09637486.2017.1353954.
- Abaturov A, Nikulina A, Nikulin D. TAS2R38 taste receptor gene and metabolically unhealthy obesity. Metabolism Clinical and Experimental. 2022;28:155003. doi: 10.1016/j.metabol.2021.155003.
- Kim HY, Choi JH. TAS2R38 bitterness receptor genetic varia–tion is associated with diet quality in Koreans. Appetite. 2024 Sep 1;200:107561. doi: 10.1016/j.appet.2024.107561.
- Wang Q, Liszt KI, Depoortere I. Extra-oral bitter taste receptors: New targets against obesity? Peptides. 2020 Feb 21;127:170284. doi: 10.1016/j.peptides.2020.170284.
- Behrens M, Lang T. Extra-Oral Taste Receptors-Function, Disease, and Perspectives. Front Nutr. 2022 Apr 4;9:881177. doi: 10.3389/fnut.2022.881177.
- Ki SY, Jeong YT. Taste Receptors beyond Taste Buds. Int J Mol Sci. 2022 Aug 26;23(17):9677. doi: 10.3390/ijms23179677.
- Harris JC, Lee RJ, Carey RM. Extragustatory bitter taste receptors in head and neck health and disease. J Mol Med (Berl). 2024 Dec;102(12):1413-1424. doi: 10.1007/s00109-024-02490-0.
- Cancello R, Micheletto G, Meta D, et al. Expanding the role of bitter taste receptor in extra oral tissues: TAS2R38 is expressed in human adipocytes. Adipocyte. 2020 Dec;9(1):7-15. doi: 10.1080/21623945.2019.1709253.